Research Topics
- Genomic Characterization of Cancer
- RNA Expression
- Nanoparticle Therapy (siRNA and small molecule)
- Personalized Medicine
Research Overview
The overarching goal of our research is to improve the outcome for patients with cancer. To do so, we seek to better understand the underlying genetic mechanisms that drive the development of cancer, or oncogenesis. In addition, we hope to utilize this newfound knowledge to identify therapeutic targets and deliver targeted therapies to tumor cells using nanoparticle delivery therapy.
The fundamental reason for our approach is based on the observation that current therapies are not specific and are generally delivered indiscriminately to normal and tumor tissue alike. In most cases, the fundamental defect in a given cancer is unknown, and there is no therapy available to address that defect. Certain tumors in young patients differ from typical adult tumors. We have some knowledge of key genetic defects, such as the universally present chimeric gene found in Ewing sarcoma, a primitive tumor of bone and soft tissue with a 50% survival rate that has not improved in over 20 years. We have, therefore, focused our attention on sarcomas in adolescents and young adults, where sarcomas constitute the single largest group of cancers.
Our approach is two-fold. We seek to:
- Understand the genetic mechanisms underlying these cancers, and once having identified the fundamental ‘driver’ genetic defects, document their biologic function and potential as therapeutic targets.
- Treat models of the disease using tumor-specified targeted nanoparticle therapy against targets identified as in 1, above, with the longer term goal of introducing these targeted therapies for the treatment of patients with these tumors.
We have been funded for over 10 years by the National Cancer Institute (NCI) to develop genomics-based biomarker profiles to predict outcomes, identify patients for risk-stratified therapies and identify potential therapeutic targets. Initially we used Affymetrix microarrays that identified only protein encoding RNA transcripts (mRNA). Later, we employed arrays that detect RNA expression across the entire genome (i.e., human exon, or HuEx, arrays) and discovered that the non-coding RNA transcripts (ncRNA) were far superior to mRNA for these purposes. More recently, as part of an initiative funded by the Department of Defense, we have turned to newer technology, based on next-generation DNA sequencing, to explore total RNA expression (RNA seq) that includes both coding and non-coding RNA expression. In the process, we have identified entirely new classes of RNA transcripts, such as the recently described vlnc RNAs. We have also used newer expression technology such as NanoString to apply the biomarker profiles we have developed to routinely available, formalin fixed, paraffin embedded specimens for use in risk stratification in future Phase III clinical trials. Ultimately, we hope to use the new knowledge generated by these exploratory studies to develop targeted therapies, employing the targeted nanoparticles we have developed over the past several years with funding from the Whittier Foundation, Hughes Foundation, and the National Science Foundation (NSF). As one example, we showed that the Ewing’s specific ncRNA, AK057037, which we identified, is also functional in this disease; when expression is ablated, the tumor becomes less invasive and metastatic in our murine model system. Similarly, when we target the EWS-FLI1 chimeric gene transcript with siRNA containing nanoparticles specific for the chimeric gene, tumor growth and metastasis is dramatically diminished, with no toxic side effects on normal tissue. Such approaches will hopefully translate to the clinic one day, fulfilling the long term goal of our research program.
Key Findings
- First to recognize that non-coding RNA expression is more biologically relevant than coding RNA expression and generates better diagnostic, prognostic and therapeutic target biomarker profiles.
- demonstrated that ncRNA transcripts are functional, playing a role in cancer and embryonic development.
- Derived fundamental models of functional networks of non-coding and coding RNA expression in cancer using a murine model of Ewing sarcoma.
- Proved that the ncRNA AK057037 is a direct functional target of the EWS-ets chimeric gene found in Ewing sarcoma.
- Demonstrated that ablation of EWS-ets expression ablates AK057037 expression and results in diminished invasion and metastasis in Ewing sarcoma.
- Discovered a new class of very large (>100,000 nucleotides) ncRNA, termed vlncRNA, that can be tumor specific and related to overall prognosis.
Non-Coding Genes Account for the Most Transcription from the Genome.
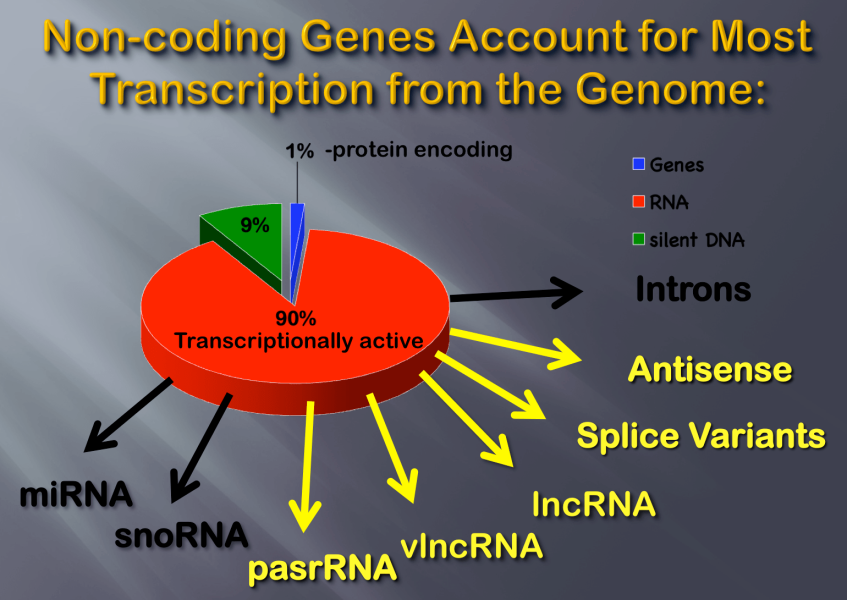
Only ~2% of the genome is used for coding gene expression and protein synthesis. Most of the rest of the genome is transcriptionally active, as illustrated. These transcripts are generically called non-coding RNA, or ncRNA, but are composed of many subtypes with discrete functions, like micro RNA, or miRNA. Many poorly understand classes are also found, like lncRNA (>100 nucleotides) and vlncRNA (>100,000 nucleotides).
Cellular Localization of ncRNA Transcript
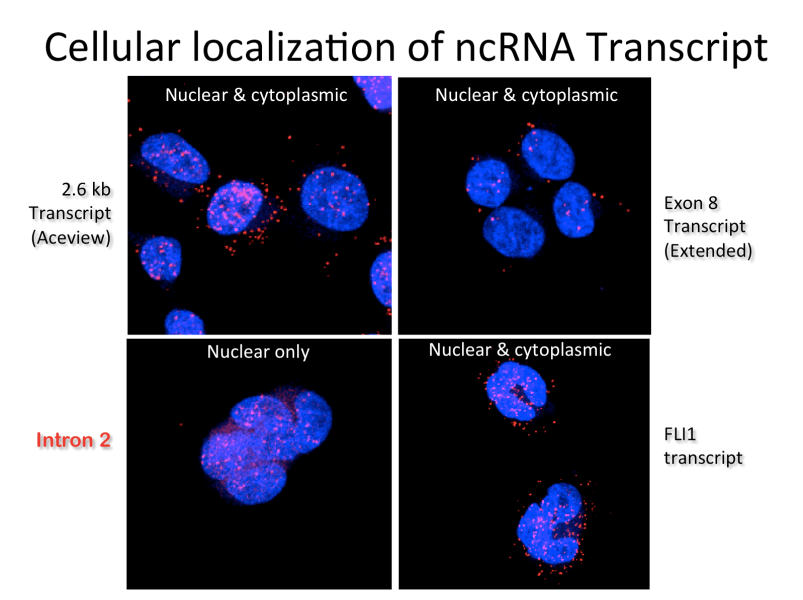
ncRNA is often predominantly, if not exclusively, retained in the nucleus, but in many cases also translates to the cytoplasm after splicing like traditional mRNA transcripts.
This is documented for a typical ncRNA by sequence-specific fluorescent in situ hybridization, or FISH, as shown here.
AK paired end RNA Sequencing
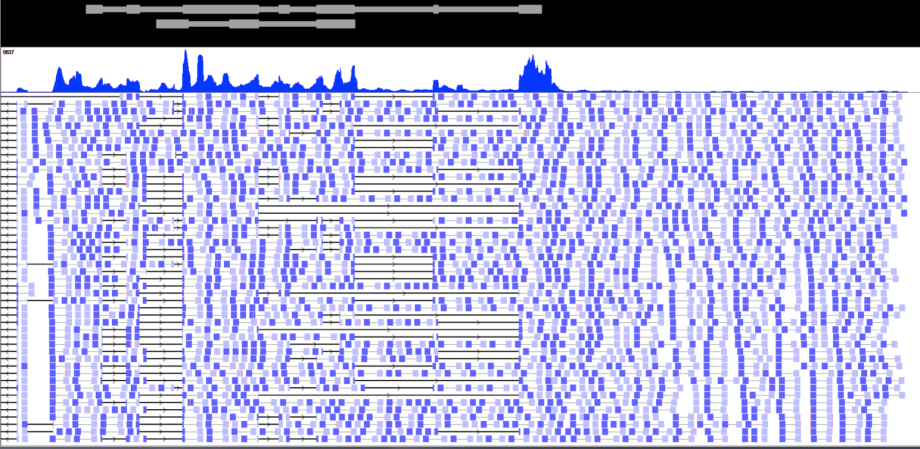
While overall expression levels are easily assessed by gene expression microarrays, the wide variety of splice isoforms found in all genes, coding and non-coding alike, is not.
This figure illustrates the wide variation in splicing as assessed by next generation RNA sequencing, or RNA seq. The upper panel shows relative abundance, akin to that measured by microarrays; the lower panel documents widespread variation in transcript structure, or isoforms, not detectable by microarrays.
Ewing's Non-coding RNA Splicing Variation in Primary vs. Metastasis
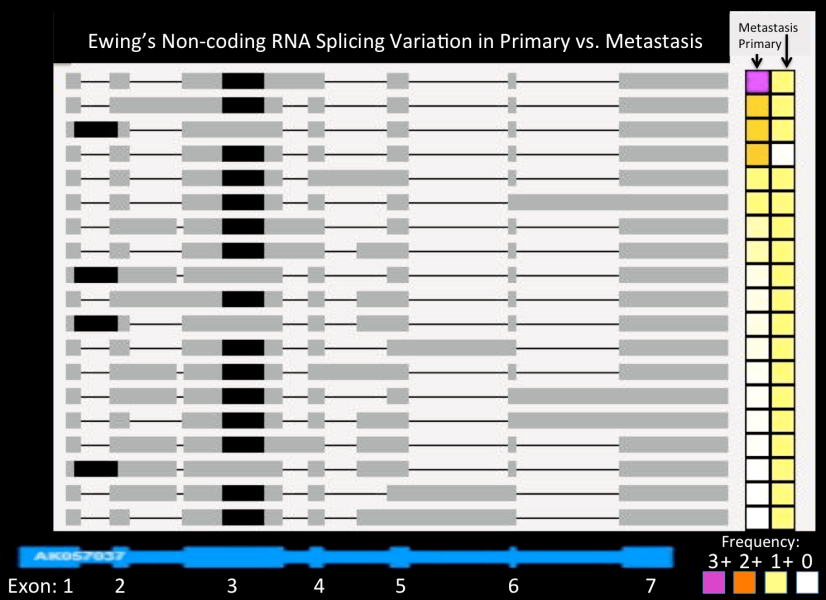
The potential importance of gene splicing is illustrated in this figure. Here, various isoforms of a Ewing-specific ncRNA, AK057037, is shown in gray, compared to the RefSeq annotated structure shown in blue at the bottom. Note that only one version corresponds to this canonical structure.
More importantly, isoform usage varies tremendously between the patient’s primary and metastatic tumor, as shown on the right in a color-coded matrix. The abundance is linked to color, from high (pink) to low (white). In the primary tumor, virtually all transcripts are accounted for by the top isoform (pink cell in the left column of the matrix). In stark contrast, there is no dominant form in the metastasis (right column, with all variants expressed at low levels, denoted by yellow cells). This suggests that the ability to control isoform use and splicing was lost in the more aggressive metastasis that resulted in the patient’s death.
Nanostring RNA Expression Values
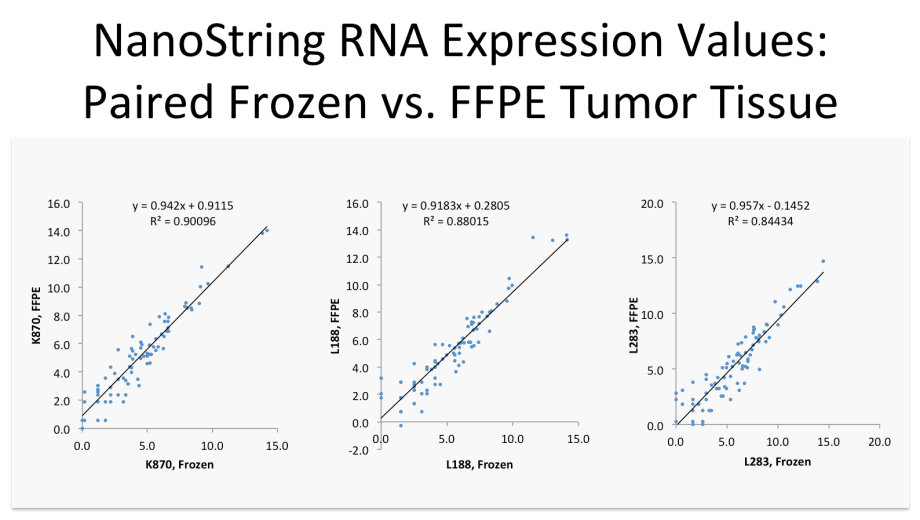
When expression levels and isoform structure and usage are understood, it is possible to utilize translational technologies like qPCR, NanoString and similar methods to measure expression values of any RNA transcript.
Here, the expression values for a 78-feature prognostic sarcoma biomarker profile are plotted for a patient's frozen tumor tissue (x axis) versus those from the same tumor tissue after fixation and embedding, on the y axis. There is a strong correlation between the two, indicating that the less expensive assay can be used on highly degraded, FFPE tumor specimens (NanoString) and will deliver RNA expression values essentially identical to those from the original ideal frozen tissue, a key requirement before such biomarkers can be used in a clinical setting where only FFPE tissue is routinely available.
Current Funding
Beckman Foundation (Ryan)
06/01/08-05/31/14
Doheny Eye Institute $200,000/yr
Macular Degeneration Research
This grant involves four PI’s at the University of Southern California and Doheny Eye Institute. Dr. Triche’s role is to provide bioinformatics development and analysis support for the other PIs, as well as gene expression profiles generated by the core microarray facility.
Role: Co-Investigator
National Cancer Institute
2 P30 CA14089-31 (Jones) 12/01/05-11/30/15
$125,000/yr
Microarray Core Facility, USC Comprehensive Cancer Center Core Grant
The major goal of this core facility located at Childrens Hospital is to provide the equipment necessary for the analysis of cooperative group pediatric cancer cases in part (50%), and the remaining capacity is being proposed for the USC Norris Comprehensive Cancer Center.
Role: Co-Investigator
Whittier Nanobiotechnology Program (Cote)
04/01/07-03/31/13
Whittier Foundation $300,000
Nanoparticle –mediated Therapeutic Targeting of Small Molecule Drugs and siRNA in Childhood Cancer
The major goal of the project is to determine whether modulation of specific tumor gene targets by siRNA, novel small molecules, or combinations thereof delivered to tumor cells in targeted 50 nm nanoparticles will maximize tumor kill and minimize systemic toxicity in a disseminated model of Ewing’s sarcomas in immunocompromised mice.
Role: Co-Principal Investigator
Department of Defense
W81XWH-07-1-0580 (Reaman)
10/01/07-09/30/12
Innovative Approaches to Therapeutic Target Identification in High-Risk Pediatric Cancers
- $1,000,000 (Year 1)
- $500,000 (Year 2)
- $500,000 (Year 3)
- $700,000 (Year 4)
The overall goal of the proposed studies is to provide a detailed, molecular map of pediatric high-risk sarcomas and distinct subtypes of acute myelogenous leukemia. The establishment of the proposed integrated and genome-wide approach to these cancers should be generalizable to other types of cancer as well as other disorders in pediatrics and adults of all ages.
Role: Co-Principal Investigator
Translation of Predictive Cancer Biomarkers Into Clinical Practice
5RC2CA148216-02 (Triche)
09/29/09-08/31/12
$1,431,000
This award supports the translation of a 24 gene metagene prognostic biomarker profile from frozen tissue to routine formalin fixed, paraffin embedded tissue. Multiple technologies to do so will be compared, the best chosen, and the best applied to over 500 FFPE tissues collected from other COG D9803 patients.
Next Generation DNA/RNA Sequencing to Define and Treat High Risk Childhood Cancers
W81XWH-10-1-0704
09/10/10-10/09/12
Department of Defense
$1,051,000 (2 Years)
This grant will allow the purchase and deployment of next gen DNA sequencing technology at Children's Hospital Los Angeles.
NCI Target
Funded Investigator through this program.


